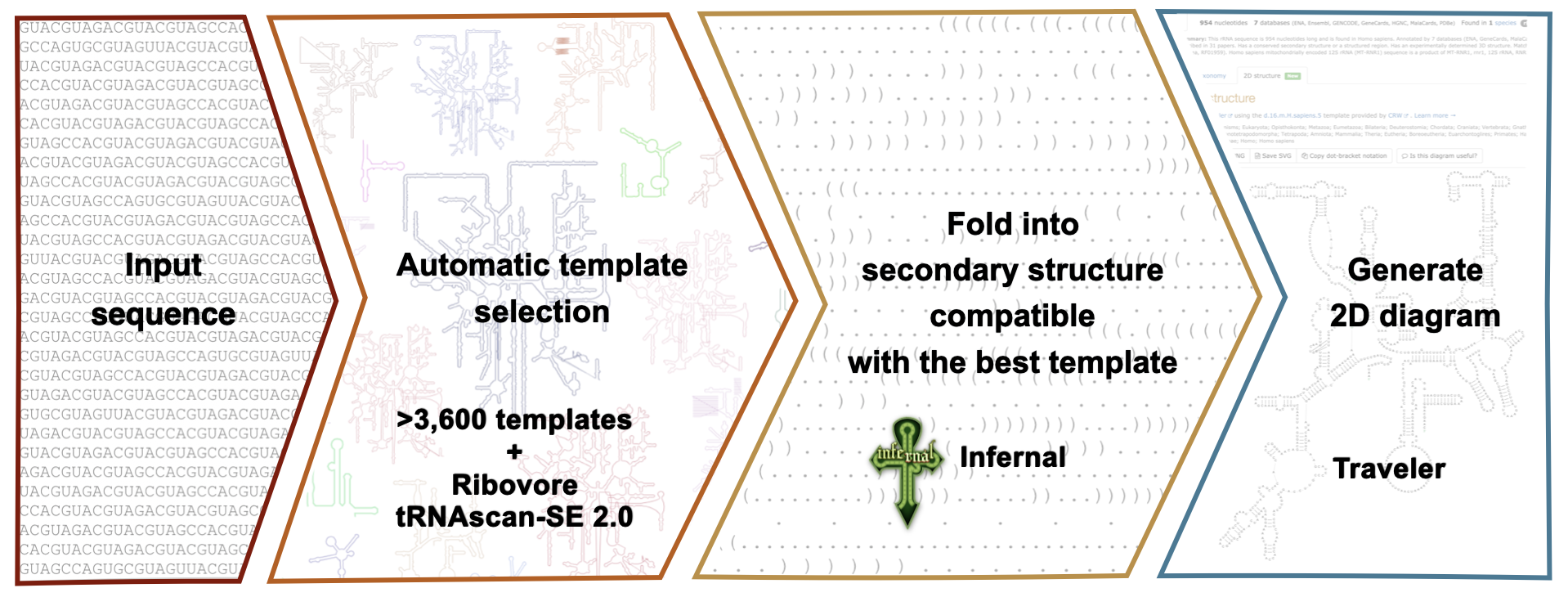
RNAcentral generates secondary structure (2D) diagrams using the R2DT software that visualises RNA structure using standard layouts or templates. Learn more about the method in the R2DT paper or check out the code on GitHub.
RNAcentral stores RNA secondary structures computed using R2DT for millions of non-coding RNAs. The structures are displayed using a library of ~4,000 templates, including:
For each sequence the R2DT pipeline automatically selects the template using ribovore, folds the sequence into the template structure using Infernal, and visualises it using Traveler.
The secondary structures are displayed in text search results, on RNA sequence pages, and in sequence similarity search results. For example, text search results include simplified outlines of the structures:
Clicking on any thumbnail shows a detailed diagram, such as the following fly RNAse P structure:
The differences between the template and the sequence are highlighted in colour. The nucleotides shown in black are identical to the template while the nucleotides that differ from the template are shown in several colours depending on whether it's an insertion (magenta) or modification (green).
A secondary structure can be generated for an RNA sequence using the R2DT web application.
By default, R2DT chooses a template automatically, but it is possible to choose a template manually by clicking Show advanced and browsing the templates. This option could be useful to highlight species-specific regions. For example, one could display the human small SSU rRNA using a bacterial template.
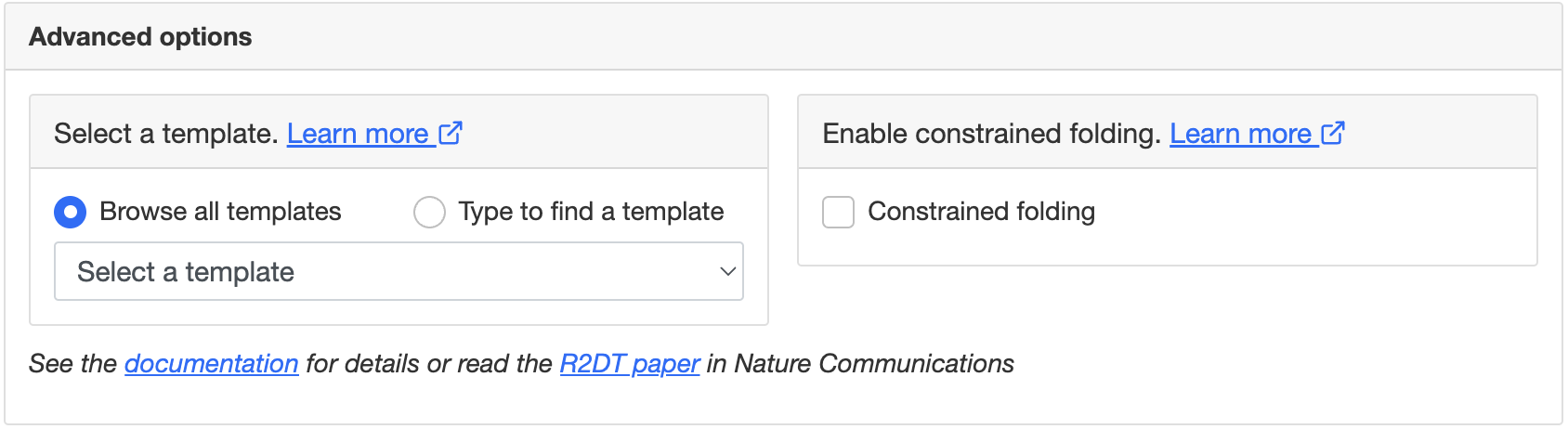
The advanced options also enable constrained folding, which uses RNAfold from the Vienna RNA package to add base pairs for the single-stranded regions that are not modelled by the templates. For example, this method can improve microRNA diagrams where the Rfam consensus structure has long dangling ends or large hairpin loops. It can also fold species-specific insertions found in many rRNAs:
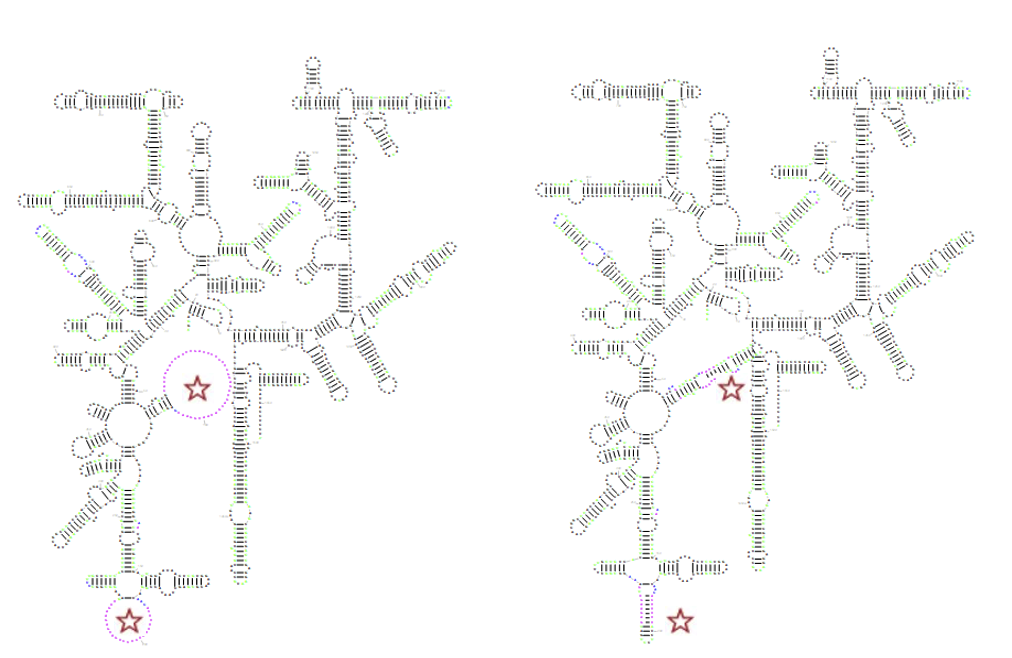
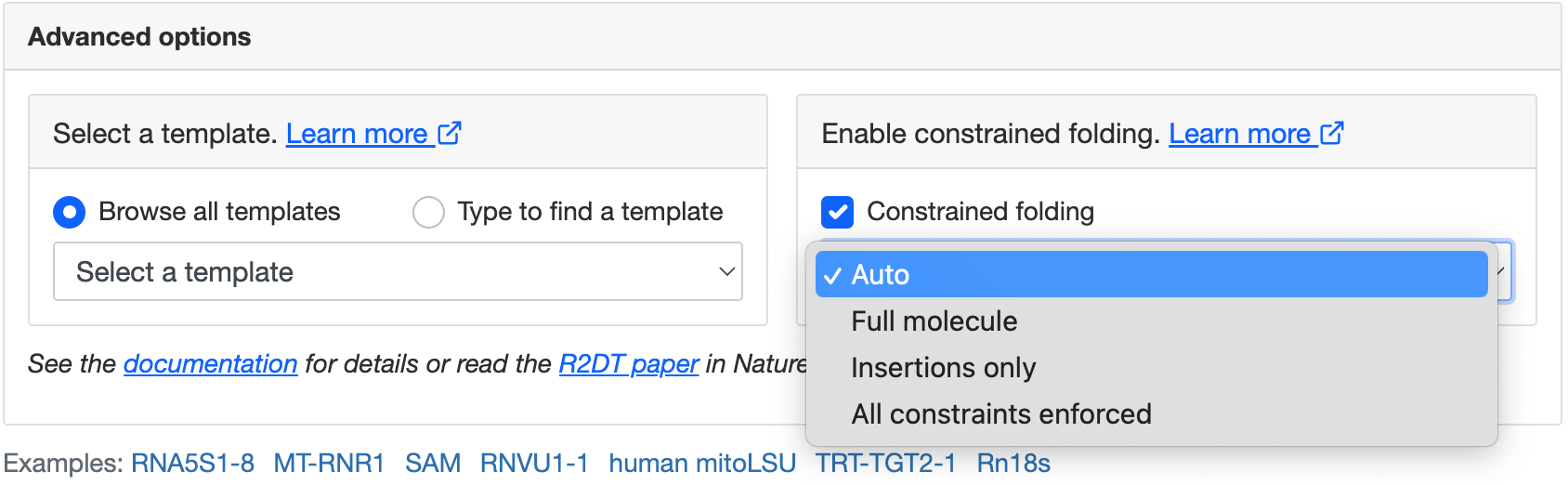
There are four ways of using constrained folding:
To use constrained folding, choose one of the modes in the advanced options:
It is possible to embed an R2DT secondary structure into any website, and it has already been added to FlyBase, SGD, and Nucleic Acid Knowledgebase.
Find out more about how to integrate this widget into your website.