If you're wondering how RNAcentral might help you, here are some ideas from papers citing RNAcentral and tweets with the #UsingRNAcentral hashtag.
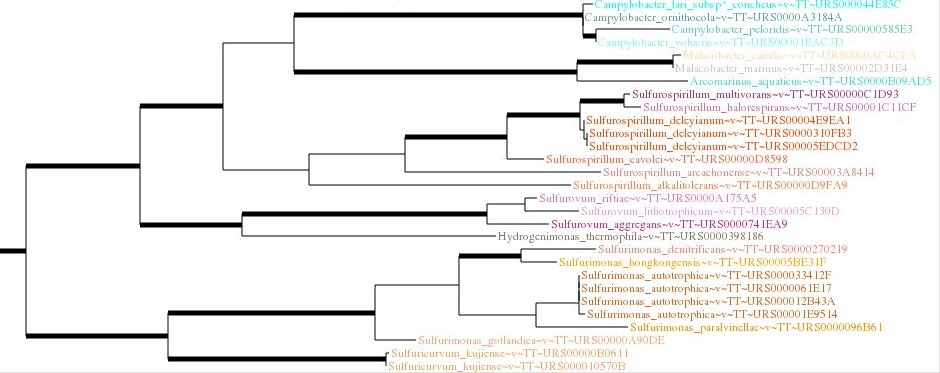
RNAcentral provides a comprehensive set of ncRNA sequences of all ncRNA types from over 780,000 species which can be used to identify new ncRNA sequences that have not been seen before or to annotate a new genome.


Each RNAcentral sequence has a stable identifier that uniquely corresponds to the RNA sequence. These identifiers are used to attach information extracted by curators from scientific literature:



RNAcentral has a powerful API that can be used to access sequences and associated metadata, such as descriptions, cross-references, or secondary structures:
